Algorithms of Evolution
Evolutionary mechanisms have inspired mathematical modeling, and computational methods underpin investigations into bioinformatics and biological evolution.
Math & Biology
Mathematics Is Biology's Next Microscope, Only Better; Biology Is Mathematics' Next Physics, Only Better
"Although mathematics has long been intertwined with the biological sciences, an explosive synergy between biology and mathematics seems poised to enrich and extend both fields greatly in the coming decades (Levin 1992; Murray 1993; Jungck 1997; Hastings et al. 2003; Palmer et al. 2003; Hastings and Palmer 2003). Biology will increasingly stimulate the creation of qualitatively new realms of mathematics. Why? In biology, ensemble properties emerge at each level of organization from the interactions of heterogeneous biological units at that level and at lower and higher levels of organization (larger and smaller physical scales, faster and slower temporal scales). New mathematics will be required to cope with these ensemble properties and with the heterogeneity of the biological units that compose ensembles at each level."
Clustered Image Map of Gene Expression–Drug Activity Correlations : Table Mathematics Arising from Biological Problems in: Joel E. Cohen, Mathematics Is Biology's Next Microscope, Only Better; Biology Is Mathematics' Next Physics, Only Better, PLOS Biology Volume 2 Issue 12 DECEMBER 2004
"Although mathematics has long been intertwined with the biological sciences, an explosive synergy between biology and mathematics seems poised to enrich and extend both fields greatly in the coming decades (Levin 1992; Murray 1993; Jungck 1997; Hastings et al. 2003; Palmer et al. 2003; Hastings and Palmer 2003). Biology will increasingly stimulate the creation of qualitatively new realms of mathematics. Why? In biology, ensemble properties emerge at each level of organization from the interactions of heterogeneous biological units at that level and at lower and higher levels of organization (larger and smaller physical scales, faster and slower temporal scales). New mathematics will be required to cope with these ensemble properties and with the heterogeneity of the biological units that compose ensembles at each level."
Clustered Image Map of Gene Expression–Drug Activity Correlations : Table Mathematics Arising from Biological Problems in: Joel E. Cohen, Mathematics Is Biology's Next Microscope, Only Better; Biology Is Mathematics' Next Physics, Only Better, PLOS Biology Volume 2 Issue 12 DECEMBER 2004
phylogenetics versus phenetics
Phylogenetic separation into evolutionary relationships (clades), based on comparison of genomes is likely to supplant traditional phenotypical (phenetic) taxonomies.
Phenetic systems group organisms based on mutual similarity of phenotypic (physical and chemical) characteristics. Such taxonomies aim to group organisms according to shared characteristics against the background of biological diversity. Phenetic groupings may or may not correlate with evolutionary relationships.
Phenetic systems group organisms based on mutual similarity of phenotypic (physical and chemical) characteristics. Such taxonomies aim to group organisms according to shared characteristics against the background of biological diversity. Phenetic groupings may or may not correlate with evolutionary relationships.
duplications of gene & genome
Modeling gene and genome duplications in eukaryotes. modified:
Recent analysis of complete eukaryotic genome sequences has revealed that gene duplication has been rampant. Moreover, next to a continuous mode of gene duplication, in many eukaryotic organisms the complete genome has been duplicated in their evolutionary past. Such large-scale gene duplication events have been associated with important evolutionary transitions or major leaps in development and adaptive radiations of species. Here, we present an evolutionary model that simulates the duplication dynamics of genes, considering genome-wide duplication events and a continuous mode of gene duplication. Modeling the evolution of the different functional categories of genes assesses the importance of different duplication events for gene families involved in specific functions or processes. By applying our model to the Arabidopsis genome, for which there is compelling evidence for three whole-genome duplications, we show that gene loss is strikingly different for large-scale and small-scale duplication events and highly biased toward certain functional classes. We provide evidence that some categories of genes were almost exclusively expanded through large-scale gene duplication events. In particular, we show that the three whole-genome duplications in Arabidopsis have been directly responsible for >90% of the increase in transcription factors, signal transducers, and developmental genes in the last 350 million years. Our evolutionary model is widely applicable and can be used to evaluate different assumptions regarding small- or large-scale gene duplication events in eukaryotic genomes.
Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. Modeling gene and genome duplications in eukaryotes. (Free Full Text Article) Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5454-9. Epub 2005 Mar 30.
The hidden duplication past of Arabidopsis thaliana. [Proc Natl Acad Sci U S A. 2002] PMID: 12374856
Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. [Genome Biol. 2006] PMID: 16507168
Structural divergence of chromosomal segments that arose from successive duplication events in the Arabidopsis genome. [Nucleic Acids Res. 2003] PMID: 12582254
Investigating ancient duplication events in the Arabidopsis genome. [J Struct Funct Genomics. 2003] PMID: 12836691
Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. [Trends Genet. 2004] PMID: 15363896
See all Related Articles...
Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity.
Controversy surrounds the apparent rising maximums of morphological complexity during eukaryotic evolution, with organisms increasing the number and nestedness of developmental areas as evidenced by morphological elaborations reflecting area boundaries. No "predictable drive" to increase this sort of complexity has been reported. Recent genetic data and theory in the general area of gene dosage effects has engendered a robust "gene balance hypothesis," with a theoretical base that makes specific predictions as to gene content changes following different types of gene duplication. Genomic data from both chordate and angiosperm genomes fit these predictions: Each type of duplication provides a one-way injection of a biased set of genes into the gene pool. Tetraploidies and balanced segments inject bias for those genes whose products are the subunits of the most complex biological machines or cascades, like transcription factors (TFs) and proteasome core proteins. Most duplicate genes are removed after tetraploidy. Genic balance is maintained by not removing those genes that are dose-sensitive, which tends to leave duplicate "functional modules" as the indirect products (spandrels) of purifying selection. Functional modules are the likely precursors of coadapted gene complexes, a unit of natural selection. The result is a predictable drive mechanism where "drive" is used rigorously, as in "meiotic drive." Rising morphological gain is expected given a supply of duplicate functional modules. All flowering plants have survived at least three large-scale duplications/diploidizations over the last 300 million years (Myr). An equivalent period of tetraploidy and body plan evolution may have ended for animals 500 million years ago (Mya). We argue that "balanced gene drive" is a sufficient explanation for the trend that the maximums of morphological complexity have gone up, and not down, in both plant and animal eukaryotic lineages.Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006 Jul;16(7):805-14.
Modeling gene and genome duplications in eukaryotes. [Proc Natl Acad Sci U S A. 2005] PMID: 15800040 Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. [Genome Res. 2006] PMID: 16760422 Widespread genome duplications throughout the history of flowering plants. [Genome Res. 2006] PMID: 16702410 New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. [Genome Res. 2003] PMID: 12799346 See all Related Articles...
Extensive genomic duplication during early chordate evolution.Opinions on the hypothesis that ancient genome duplications contributed to the vertebrate genome range from strong skepticism to strong credence. Previous studies concentrated on small numbers of gene families or chromosomal regions that might not have been representative of the whole genome, or used subjective methods to identify paralogous genes and regions. Here we report a systematic and objective analysis of the draft human genome sequence to identify paralogous chromosomal regions (paralogons) formed during chordate evolution and to estimate the ages of duplicate genes. We found that the human genome contains many more paralogons than would be expected by chance. Molecular clock analysis of all protein families in humans that have orthologs in the fly and nematode indicated that a burst of gene duplication activity took place in the period 350 650 Myr ago and that many of the duplicate genes formed at this time are located within paralogons. Our results support the contention that many of the gene families in vertebrates were formed or expanded by large-scale DNA duplications in an early chordate. Considering the incompleteness of the sequence data and the antiquity of the event, the results are compatible with at least one round of polyploidy.
McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002 Jun;31(2):200-4. Epub 2002 May 28. Comment in: Nat Genet. 2002 Jun;31(2):128-9.
Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. [Mol Biol Evol. 2004] PMID: 15014147 New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. [Genome Res. 2003] PMID: 12799346 Ancient large-scale genome duplications: phylogenetic and linkage analyses shed light on chordate genome evolution. [Mol Biol Evol. 1998] PMID: 9729879 Phylogenetic analyses alone are insufficient to determine whether genome duplication(s) occurred during early vertebrate evolution. [J Exp Zoolog B Mol Dev Evol. 2003] PMID: 14508816 Phylogenetic analysis of T-Box genes demonstrates the importance of amphioxus for understanding evolution of the vertebrate genome. [Genetics. 2000] PMID: 11063699See all Related Articles...
Timing and mechanism of ancient vertebrate genome duplications -- the adventure of a hypothesis.Complete genome doubling has long-term consequences for the genome structure and the subsequent evolution of an organism. It has been suggested that two genome duplications occurred at the origin of vertebrates (known as the 2R hypothesis). However, there has been considerable debate as to whether these were two successive duplications, or whether a single duplication occurred, followed by large-scale segmental duplications. In this article, we review and compare the evidence for the 2R duplications from vertebrate genomes with similar data from other more recent polyploids.
Panopoulou G, Poustka AJ. Timing and mechanism of ancient vertebrate genome duplications -- the adventure of a hypothesis. Trends Genet. 2005 Oct;21(10):559-67
Evolution and diversity of fish genomes. [Curr Opin Genet Dev. 2003] PMID: 14638319
Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. [Proc Natl Acad Sci U S A. 2004] PMID: 14757817Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. [Mol Biol Evol. 2004] PMID: 15014147 Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. [Nature. 2004] PMID: 15496914 See all Related Articles...
Turning the clock back on ancient genome duplication. [Curr Opin Genet Dev. 2003] PMID: 14638327 Were vertebrates octoploid? [Philos Trans R Soc Lond B Biol Sci. 2002] PMID: 12028790 Phylogenetic dating and characterization of gene duplications in vertebrates: the cartilaginous fish reference. [Mol Biol Evol. 2004] PMID: 14694077 Phylogenetic analyses alone are insufficient to determine whether genome duplication(s) occurred during early vertebrate evolution. [J Exp Zoolog B Mol Dev Evol. 2003] PMID: 14508816 Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. [Mol Biol Evol. 2002] PMID: 12200472See all Related Articles...
~ Basic mechanisms of evolution ~ Duplication ^ Population Genetics Tables Mechanisms of Biological Evolution : Gene Regulation in E.coli :
Recent analysis of complete eukaryotic genome sequences has revealed that gene duplication has been rampant. Moreover, next to a continuous mode of gene duplication, in many eukaryotic organisms the complete genome has been duplicated in their evolutionary past. Such large-scale gene duplication events have been associated with important evolutionary transitions or major leaps in development and adaptive radiations of species. Here, we present an evolutionary model that simulates the duplication dynamics of genes, considering genome-wide duplication events and a continuous mode of gene duplication. Modeling the evolution of the different functional categories of genes assesses the importance of different duplication events for gene families involved in specific functions or processes. By applying our model to the Arabidopsis genome, for which there is compelling evidence for three whole-genome duplications, we show that gene loss is strikingly different for large-scale and small-scale duplication events and highly biased toward certain functional classes. We provide evidence that some categories of genes were almost exclusively expanded through large-scale gene duplication events. In particular, we show that the three whole-genome duplications in Arabidopsis have been directly responsible for >90% of the increase in transcription factors, signal transducers, and developmental genes in the last 350 million years. Our evolutionary model is widely applicable and can be used to evaluate different assumptions regarding small- or large-scale gene duplication events in eukaryotic genomes.
Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. Modeling gene and genome duplications in eukaryotes. (Free Full Text Article) Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5454-9. Epub 2005 Mar 30.
The hidden duplication past of Arabidopsis thaliana. [Proc Natl Acad Sci U S A. 2002] PMID: 12374856
Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. [Genome Biol. 2006] PMID: 16507168
Structural divergence of chromosomal segments that arose from successive duplication events in the Arabidopsis genome. [Nucleic Acids Res. 2003] PMID: 12582254
Investigating ancient duplication events in the Arabidopsis genome. [J Struct Funct Genomics. 2003] PMID: 12836691
Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. [Trends Genet. 2004] PMID: 15363896
See all Related Articles...
Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity.
Controversy surrounds the apparent rising maximums of morphological complexity during eukaryotic evolution, with organisms increasing the number and nestedness of developmental areas as evidenced by morphological elaborations reflecting area boundaries. No "predictable drive" to increase this sort of complexity has been reported. Recent genetic data and theory in the general area of gene dosage effects has engendered a robust "gene balance hypothesis," with a theoretical base that makes specific predictions as to gene content changes following different types of gene duplication. Genomic data from both chordate and angiosperm genomes fit these predictions: Each type of duplication provides a one-way injection of a biased set of genes into the gene pool. Tetraploidies and balanced segments inject bias for those genes whose products are the subunits of the most complex biological machines or cascades, like transcription factors (TFs) and proteasome core proteins. Most duplicate genes are removed after tetraploidy. Genic balance is maintained by not removing those genes that are dose-sensitive, which tends to leave duplicate "functional modules" as the indirect products (spandrels) of purifying selection. Functional modules are the likely precursors of coadapted gene complexes, a unit of natural selection. The result is a predictable drive mechanism where "drive" is used rigorously, as in "meiotic drive." Rising morphological gain is expected given a supply of duplicate functional modules. All flowering plants have survived at least three large-scale duplications/diploidizations over the last 300 million years (Myr). An equivalent period of tetraploidy and body plan evolution may have ended for animals 500 million years ago (Mya). We argue that "balanced gene drive" is a sufficient explanation for the trend that the maximums of morphological complexity have gone up, and not down, in both plant and animal eukaryotic lineages.Freeling M, Thomas BC. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006 Jul;16(7):805-14.
Modeling gene and genome duplications in eukaryotes. [Proc Natl Acad Sci U S A. 2005] PMID: 15800040 Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. [Genome Res. 2006] PMID: 16760422 Widespread genome duplications throughout the history of flowering plants. [Genome Res. 2006] PMID: 16702410 New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. [Genome Res. 2003] PMID: 12799346 See all Related Articles...
Extensive genomic duplication during early chordate evolution.Opinions on the hypothesis that ancient genome duplications contributed to the vertebrate genome range from strong skepticism to strong credence. Previous studies concentrated on small numbers of gene families or chromosomal regions that might not have been representative of the whole genome, or used subjective methods to identify paralogous genes and regions. Here we report a systematic and objective analysis of the draft human genome sequence to identify paralogous chromosomal regions (paralogons) formed during chordate evolution and to estimate the ages of duplicate genes. We found that the human genome contains many more paralogons than would be expected by chance. Molecular clock analysis of all protein families in humans that have orthologs in the fly and nematode indicated that a burst of gene duplication activity took place in the period 350 650 Myr ago and that many of the duplicate genes formed at this time are located within paralogons. Our results support the contention that many of the gene families in vertebrates were formed or expanded by large-scale DNA duplications in an early chordate. Considering the incompleteness of the sequence data and the antiquity of the event, the results are compatible with at least one round of polyploidy.
McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002 Jun;31(2):200-4. Epub 2002 May 28. Comment in: Nat Genet. 2002 Jun;31(2):128-9.
Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. [Mol Biol Evol. 2004] PMID: 15014147 New evidence for genome-wide duplications at the origin of vertebrates using an amphioxus gene set and completed animal genomes. [Genome Res. 2003] PMID: 12799346 Ancient large-scale genome duplications: phylogenetic and linkage analyses shed light on chordate genome evolution. [Mol Biol Evol. 1998] PMID: 9729879 Phylogenetic analyses alone are insufficient to determine whether genome duplication(s) occurred during early vertebrate evolution. [J Exp Zoolog B Mol Dev Evol. 2003] PMID: 14508816 Phylogenetic analysis of T-Box genes demonstrates the importance of amphioxus for understanding evolution of the vertebrate genome. [Genetics. 2000] PMID: 11063699See all Related Articles...
Timing and mechanism of ancient vertebrate genome duplications -- the adventure of a hypothesis.Complete genome doubling has long-term consequences for the genome structure and the subsequent evolution of an organism. It has been suggested that two genome duplications occurred at the origin of vertebrates (known as the 2R hypothesis). However, there has been considerable debate as to whether these were two successive duplications, or whether a single duplication occurred, followed by large-scale segmental duplications. In this article, we review and compare the evidence for the 2R duplications from vertebrate genomes with similar data from other more recent polyploids.
Panopoulou G, Poustka AJ. Timing and mechanism of ancient vertebrate genome duplications -- the adventure of a hypothesis. Trends Genet. 2005 Oct;21(10):559-67
Evolution and diversity of fish genomes. [Curr Opin Genet Dev. 2003] PMID: 14638319
Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray-finned fishes and land vertebrates. [Proc Natl Acad Sci U S A. 2004] PMID: 14757817Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. [Mol Biol Evol. 2004] PMID: 15014147 Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. [Nature. 2004] PMID: 15496914 See all Related Articles...
Turning the clock back on ancient genome duplication. [Curr Opin Genet Dev. 2003] PMID: 14638327 Were vertebrates octoploid? [Philos Trans R Soc Lond B Biol Sci. 2002] PMID: 12028790 Phylogenetic dating and characterization of gene duplications in vertebrates: the cartilaginous fish reference. [Mol Biol Evol. 2004] PMID: 14694077 Phylogenetic analyses alone are insufficient to determine whether genome duplication(s) occurred during early vertebrate evolution. [J Exp Zoolog B Mol Dev Evol. 2003] PMID: 14508816 Analysis of lamprey and hagfish genes reveals a complex history of gene duplications during early vertebrate evolution. [Mol Biol Evol. 2002] PMID: 12200472See all Related Articles...
~ Basic mechanisms of evolution ~ Duplication ^ Population Genetics Tables Mechanisms of Biological Evolution : Gene Regulation in E.coli :
epistasis
Epistasis is defined as the influence of the genotype at one locus on the effect of a mutation at another locus. Thus, epistasis is the interaction between two or more genes to control a single phenotype.
Epistasis plays a crucial role in a variety of evolutionary phenomena such as speciation, population bottle necks, and the evolution of genetic architecture – the evolution of dominance, canalization, and genetic correlations.
Crossing dihybrids produces a modified Mendelian ratio – a 9:3:3:1 ratio for two dihybrids, in accordance with Mendel's second law, the law of independent assortment.
Genetic measurement of theory of epistatic effects.
Epistasis is defined as the influence of the genotype at one locus on the effect of a mutation at another locus. As such it plays a crucial role in a variety of evolutionary phenomena such as speciation, population bottle necks, and the evolution of genetic architecture (i.e., the evolution of dominance, canalization, and genetic correlations). In mathematical population genetics, however, epistasis is often represented as a mere noise term in an additive model of gene effects. In this paper it is argued that epistasis needs to be scaled in a way that is more directly related to the mechanisms of evolutionary change. A review of general measurement theory shows that the scaling of a quantitative concept has to reflect the empirical relationships among the objects. To apply these ideas to epistatic mutation effects, it is proposed to scale A x A epistatic effects as the change in the magnitude of the additive effect of a mutation at one locus due to a mutation at a second locus. It is shown that the absolute change in the additive effect at locus A due to a substitution at locus B is always identical to the absolute change in B due to the substitution at A. The absolute A x A epistatic effects of A on B and of B on A are identical, even if the relative effects can be different. The proposed scaling of A x A epistasis leads to particularly simple equations for the decomposition of genotypic variance. The Kacser Burns model of metabolic flux is analyzed for the presence of epistatic effects on flux. It is shown that the non-linearity of the Kacser Burns model is not sufficient to cause A x A epistasis among the genes coding for enzymes. It is concluded that non-linearity of the genotype-phenotype map is not sufficient to cause epistasis. Finally, it is shown that there exist correlations among the additive and epistatic effects among pairs of loci, caused by the inherent symmetries of Mendelian genetic systems. For instance, it is shown that a mutation that has a larger than average additive effect will tend to decrease the additive effect of a second mutation, i.e., it will tend to have a negative (canalizing) interaction with a subsequent gene substitution. This is confirmed in a preliminary analysis of QTL-data for adult body weight in mice.
Wagner GP, Laubichler MD, Bagheri-Chaichian H. Genetic measurement of theory of epistatic effects.. Genetica. 1998;102-103(1-6):569-80.
Epistasis plays a crucial role in a variety of evolutionary phenomena such as speciation, population bottle necks, and the evolution of genetic architecture – the evolution of dominance, canalization, and genetic correlations.
Crossing dihybrids produces a modified Mendelian ratio – a 9:3:3:1 ratio for two dihybrids, in accordance with Mendel's second law, the law of independent assortment.
Genetic measurement of theory of epistatic effects.
Epistasis is defined as the influence of the genotype at one locus on the effect of a mutation at another locus. As such it plays a crucial role in a variety of evolutionary phenomena such as speciation, population bottle necks, and the evolution of genetic architecture (i.e., the evolution of dominance, canalization, and genetic correlations). In mathematical population genetics, however, epistasis is often represented as a mere noise term in an additive model of gene effects. In this paper it is argued that epistasis needs to be scaled in a way that is more directly related to the mechanisms of evolutionary change. A review of general measurement theory shows that the scaling of a quantitative concept has to reflect the empirical relationships among the objects. To apply these ideas to epistatic mutation effects, it is proposed to scale A x A epistatic effects as the change in the magnitude of the additive effect of a mutation at one locus due to a mutation at a second locus. It is shown that the absolute change in the additive effect at locus A due to a substitution at locus B is always identical to the absolute change in B due to the substitution at A. The absolute A x A epistatic effects of A on B and of B on A are identical, even if the relative effects can be different. The proposed scaling of A x A epistasis leads to particularly simple equations for the decomposition of genotypic variance. The Kacser Burns model of metabolic flux is analyzed for the presence of epistatic effects on flux. It is shown that the non-linearity of the Kacser Burns model is not sufficient to cause A x A epistasis among the genes coding for enzymes. It is concluded that non-linearity of the genotype-phenotype map is not sufficient to cause epistasis. Finally, it is shown that there exist correlations among the additive and epistatic effects among pairs of loci, caused by the inherent symmetries of Mendelian genetic systems. For instance, it is shown that a mutation that has a larger than average additive effect will tend to decrease the additive effect of a second mutation, i.e., it will tend to have a negative (canalizing) interaction with a subsequent gene substitution. This is confirmed in a preliminary analysis of QTL-data for adult body weight in mice.
Wagner GP, Laubichler MD, Bagheri-Chaichian H. Genetic measurement of theory of epistatic effects.. Genetica. 1998;102-103(1-6):569-80.
directional epistasis
The role of epistatic gene interactions in the response to selection and the evolution of evolvability.
It has been argued that the architecture of the genotype-phenotype map determines evolvability, but few studies have attempted to quantify these effects. In this article we use the multilinear epistatic model to study the effects of different forms of epistasis on the response to directional selection. We derive an analytical prediction for the change in the additive genetic variance, and use individual-based simulations to understand the dynamics of evolvability and the evolution of genetic architecture. This shows that the major determinant for the evolution of the additive variance, and thus the evolvability, is directional epistasis. Positive directional epistasis leads to an acceleration of evolvability, while negative directional epistasis leads to canalization. In contrast, pure non-directional epistasis has little effect on the response to selection. One consequence of this is that the classical epistatic variance components, which do not distinguish directional and non-directional effects, are useless as predictors of evolutionary dynamics. The build-up of linkage disequilibrium also has negligible effects. We argue that directional epistasis is likely to have major effects on evolutionary dynamics and should be the focus of empirical studies of epistasis.
Carter AJ, Hermisson J, Hansen TF. The role of epistatic gene interactions in the response to selection and the evolution of evolvability. Theor Popul Biol. 2005 Nov;68(3):179-96. Epub 2005 Aug 24.
It has been argued that the architecture of the genotype-phenotype map determines evolvability, but few studies have attempted to quantify these effects. In this article we use the multilinear epistatic model to study the effects of different forms of epistasis on the response to directional selection. We derive an analytical prediction for the change in the additive genetic variance, and use individual-based simulations to understand the dynamics of evolvability and the evolution of genetic architecture. This shows that the major determinant for the evolution of the additive variance, and thus the evolvability, is directional epistasis. Positive directional epistasis leads to an acceleration of evolvability, while negative directional epistasis leads to canalization. In contrast, pure non-directional epistasis has little effect on the response to selection. One consequence of this is that the classical epistatic variance components, which do not distinguish directional and non-directional effects, are useless as predictors of evolutionary dynamics. The build-up of linkage disequilibrium also has negligible effects. We argue that directional epistasis is likely to have major effects on evolutionary dynamics and should be the focus of empirical studies of epistasis.
Carter AJ, Hermisson J, Hansen TF. The role of epistatic gene interactions in the response to selection and the evolution of evolvability. Theor Popul Biol. 2005 Nov;68(3):179-96. Epub 2005 Aug 24.
evolutionary hotspots in genome
Bioinformatics Window on Evolution :: Bio-IT World: "An international team including researchers from the National Human Genome Research Institute (NHGRI) has discovered that mammalian chromosomes have evolved over tens of millions of years by breaking at specific sites rather than randomly, as had been widely thought.
Writing in last week's Science, the researchers compared the genomic architecture of eight mammals: human, mouse, rat, cow, pig, dog, cat and horse. Bioinformatics analysis revealed that during evolution, the chromosomes are rearranged by breaking typically in specific locations, rather than in a random fashion, as had been widely thought. "This study shows the tremendous power of using multi-species genome comparisons to understand evolutionary processes, including those with potential relevance to human disease," comments Eric Green, NHGRI Scientific Director.
Inherited chromosomal translocations play a key role in evolution by rearranging genetic material, which in rare cases can be beneficial. They can also be deleterious - translocations are often associated with cancer. The researchers find that the chromosome breaks linked to cancer are more likely to occur in proximity to the evolutionary breakage hotspots. The authors also conclude, based on computer-generated reconstructions of the genomes of long-extinct mammals, that there was a sharp increase in the rate of chromosomal evolution among mammals following the demise of the dinosaurs some 65 million years ago"
Murphy, W.J. et al. “Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps.” Science 309, 613-617 (2005).
Writing in last week's Science, the researchers compared the genomic architecture of eight mammals: human, mouse, rat, cow, pig, dog, cat and horse. Bioinformatics analysis revealed that during evolution, the chromosomes are rearranged by breaking typically in specific locations, rather than in a random fashion, as had been widely thought. "This study shows the tremendous power of using multi-species genome comparisons to understand evolutionary processes, including those with potential relevance to human disease," comments Eric Green, NHGRI Scientific Director.
Inherited chromosomal translocations play a key role in evolution by rearranging genetic material, which in rare cases can be beneficial. They can also be deleterious - translocations are often associated with cancer. The researchers find that the chromosome breaks linked to cancer are more likely to occur in proximity to the evolutionary breakage hotspots. The authors also conclude, based on computer-generated reconstructions of the genomes of long-extinct mammals, that there was a sharp increase in the rate of chromosomal evolution among mammals following the demise of the dinosaurs some 65 million years ago"
Murphy, W.J. et al. “Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps.” Science 309, 613-617 (2005).
lateral gene transfer
Also termed horizontal gene transfer, lateral gene transfer refers to the transmission of genetic material from one organism to another organism that is not its offspring. This is compared to vertical gene transfer in which genetic material is passed from parental organisms to descendent organisms.
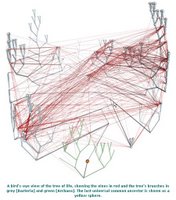 Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (phylogeny) of prokaryotes (left, adapted - click to enlarge), and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. (see The net of life: Reconstructing the microbial phylogenetic network V. Kunin, L. Goldovsky, N. Darzentas, and C. A. Ouzounis Genome Res. 1 July 2005. pdf)
Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (phylogeny) of prokaryotes (left, adapted - click to enlarge), and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. (see The net of life: Reconstructing the microbial phylogenetic network V. Kunin, L. Goldovsky, N. Darzentas, and C. A. Ouzounis Genome Res. 1 July 2005. pdf)
Three mechanisms of horizontal (lateral) gene transfer are recognized in bacteria: direct bacterial conjugation, bacteriophage mediated transduction between bacteria, and bacterial transformation by uptake and incorporation of DNA fragments. The agents of delivery in lateral gene transfer in prokaryotes are other bacteria in conjugation, viruses in transduction, and the environment in transformation.
A major form of vertical gene transfer followed serial endosymbiotic events, in which ingested purple bacteria and Cyanobacteria became eukaryotic mitochondria and chloroplasts respectively. The ingested prokaryotes are believed to have relinquished certain genes to the nuclei of their host cells, a process known as endosymbiotic gene transfer.
Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement.
When organismal phylogenies based on sequences of single marker genes are poorly resolved, a logical approach is to add more markers, on the assumption that weak but congruent phylogenetic signal will be reinforced in such multigene trees. Such approaches are valid
only when the several markers indeed have identical phylogenies, an issue which many multigene methods (such as the use of concatenated gene sequences or the assembly of supertrees) do not directly address. Indeed, even when the true history is a mixture of vertical descent for some genes and lateral gene transfer (LGT) for others, such methods produce unique topologies.
Dave MacLeod, Robert L Charlebois, Ford Doolittle and Eric Bapteste. Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement (Full Text pdf) BMC Evolutionary Biology 2005, 5:27
 Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (phylogeny) of prokaryotes (left, adapted - click to enlarge), and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. (see The net of life: Reconstructing the microbial phylogenetic network V. Kunin, L. Goldovsky, N. Darzentas, and C. A. Ouzounis Genome Res. 1 July 2005. pdf)
Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (phylogeny) of prokaryotes (left, adapted - click to enlarge), and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. (see The net of life: Reconstructing the microbial phylogenetic network V. Kunin, L. Goldovsky, N. Darzentas, and C. A. Ouzounis Genome Res. 1 July 2005. pdf)Three mechanisms of horizontal (lateral) gene transfer are recognized in bacteria: direct bacterial conjugation, bacteriophage mediated transduction between bacteria, and bacterial transformation by uptake and incorporation of DNA fragments. The agents of delivery in lateral gene transfer in prokaryotes are other bacteria in conjugation, viruses in transduction, and the environment in transformation.
A major form of vertical gene transfer followed serial endosymbiotic events, in which ingested purple bacteria and Cyanobacteria became eukaryotic mitochondria and chloroplasts respectively. The ingested prokaryotes are believed to have relinquished certain genes to the nuclei of their host cells, a process known as endosymbiotic gene transfer.
Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement.
When organismal phylogenies based on sequences of single marker genes are poorly resolved, a logical approach is to add more markers, on the assumption that weak but congruent phylogenetic signal will be reinforced in such multigene trees. Such approaches are valid
only when the several markers indeed have identical phylogenies, an issue which many multigene methods (such as the use of concatenated gene sequences or the assembly of supertrees) do not directly address. Indeed, even when the true history is a mixture of vertical descent for some genes and lateral gene transfer (LGT) for others, such methods produce unique topologies.
Dave MacLeod, Robert L Charlebois, Ford Doolittle and Eric Bapteste. Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement (Full Text pdf) BMC Evolutionary Biology 2005, 5:27
long branch attraction
Long branch attraction (LBA) is a problem in phylogenetic analyses, particularly in those analyses employing the non-parametric statistical method termed maximum parsimony.
In LBA, rapidly evolving lineages are inferred to be closely related, regardless of their true evolutionary relationships. This problem in analysis arises when the DNA of two (or more) lineages evolves rapidly. Because there are only four possible nucleotides, high rates of DNA substitution create the probability that two separate lineages will convergently evolve the same nucleotide at the same locus. In such cases, parsimony erroneously interprets this similarity as a synapomorphy, that is, as having evolved once in the common ancestor of the two lineages. The problem of LBA can be minimized by applying statistical methods that incorporate differential rates of substitution among lineages, such as maximum likelihood, or by breaking up long branches by adding taxa that are related to those with the long branches.
In LBA, rapidly evolving lineages are inferred to be closely related, regardless of their true evolutionary relationships. This problem in analysis arises when the DNA of two (or more) lineages evolves rapidly. Because there are only four possible nucleotides, high rates of DNA substitution create the probability that two separate lineages will convergently evolve the same nucleotide at the same locus. In such cases, parsimony erroneously interprets this similarity as a synapomorphy, that is, as having evolved once in the common ancestor of the two lineages. The problem of LBA can be minimized by applying statistical methods that incorporate differential rates of substitution among lineages, such as maximum likelihood, or by breaking up long branches by adding taxa that are related to those with the long branches.








































