lateral gene transfer
Also termed horizontal gene transfer, lateral gene transfer refers to the transmission of genetic material from one organism to another organism that is not its offspring. This is compared to vertical gene transfer in which genetic material is passed from parental organisms to descendent organisms.
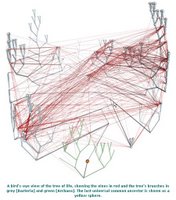 Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (phylogeny) of prokaryotes (left, adapted - click to enlarge), and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. (see The net of life: Reconstructing the microbial phylogenetic network V. Kunin, L. Goldovsky, N. Darzentas, and C. A. Ouzounis Genome Res. 1 July 2005. pdf)
Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (phylogeny) of prokaryotes (left, adapted - click to enlarge), and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. (see The net of life: Reconstructing the microbial phylogenetic network V. Kunin, L. Goldovsky, N. Darzentas, and C. A. Ouzounis Genome Res. 1 July 2005. pdf)
Three mechanisms of horizontal (lateral) gene transfer are recognized in bacteria: direct bacterial conjugation, bacteriophage mediated transduction between bacteria, and bacterial transformation by uptake and incorporation of DNA fragments. The agents of delivery in lateral gene transfer in prokaryotes are other bacteria in conjugation, viruses in transduction, and the environment in transformation.
A major form of vertical gene transfer followed serial endosymbiotic events, in which ingested purple bacteria and Cyanobacteria became eukaryotic mitochondria and chloroplasts respectively. The ingested prokaryotes are believed to have relinquished certain genes to the nuclei of their host cells, a process known as endosymbiotic gene transfer.
Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement.
When organismal phylogenies based on sequences of single marker genes are poorly resolved, a logical approach is to add more markers, on the assumption that weak but congruent phylogenetic signal will be reinforced in such multigene trees. Such approaches are valid
only when the several markers indeed have identical phylogenies, an issue which many multigene methods (such as the use of concatenated gene sequences or the assembly of supertrees) do not directly address. Indeed, even when the true history is a mixture of vertical descent for some genes and lateral gene transfer (LGT) for others, such methods produce unique topologies.
Dave MacLeod, Robert L Charlebois, Ford Doolittle and Eric Bapteste. Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement (Full Text pdf) BMC Evolutionary Biology 2005, 5:27
 Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (phylogeny) of prokaryotes (left, adapted - click to enlarge), and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. (see The net of life: Reconstructing the microbial phylogenetic network V. Kunin, L. Goldovsky, N. Darzentas, and C. A. Ouzounis Genome Res. 1 July 2005. pdf)
Horizontal gene transfer - gene swapping - has blurred the evolutionary relationships (phylogeny) of prokaryotes (left, adapted - click to enlarge), and continues to provide a mechanism for the sharing of antibiotic resistance between bacteria. (see The net of life: Reconstructing the microbial phylogenetic network V. Kunin, L. Goldovsky, N. Darzentas, and C. A. Ouzounis Genome Res. 1 July 2005. pdf)Three mechanisms of horizontal (lateral) gene transfer are recognized in bacteria: direct bacterial conjugation, bacteriophage mediated transduction between bacteria, and bacterial transformation by uptake and incorporation of DNA fragments. The agents of delivery in lateral gene transfer in prokaryotes are other bacteria in conjugation, viruses in transduction, and the environment in transformation.
A major form of vertical gene transfer followed serial endosymbiotic events, in which ingested purple bacteria and Cyanobacteria became eukaryotic mitochondria and chloroplasts respectively. The ingested prokaryotes are believed to have relinquished certain genes to the nuclei of their host cells, a process known as endosymbiotic gene transfer.
Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement.
When organismal phylogenies based on sequences of single marker genes are poorly resolved, a logical approach is to add more markers, on the assumption that weak but congruent phylogenetic signal will be reinforced in such multigene trees. Such approaches are valid
only when the several markers indeed have identical phylogenies, an issue which many multigene methods (such as the use of concatenated gene sequences or the assembly of supertrees) do not directly address. Indeed, even when the true history is a mixture of vertical descent for some genes and lateral gene transfer (LGT) for others, such methods produce unique topologies.
Dave MacLeod, Robert L Charlebois, Ford Doolittle and Eric Bapteste. Deduction of probable events of lateral gene transfer through comparison of phylogenetic trees by recursive consolidation and rearrangement (Full Text pdf) BMC Evolutionary Biology 2005, 5:27









































0 Comments:
Post a Comment
<< Home